PyRASA - Spectral parameterization in python based on IRASA#

PyRASA is a Python library designed to separate and parametrize aperiodic (fractal) and periodic (oscillatory) components in time series data based on the IRASA algorithm (Wen & Liu, 2016).
Features#
Aperiodic and Periodic Decomposition: Utilize the IRASA algorithm to decompose power spectra into aperiodic and periodic components, enabling better interpretation of neurophysiological signals.
Time Resolved Spectral Parametrization: Perform time resolved spectral parametrization, allowing you to track changes in spectral components over time.
Support for Raw and Epoched MNE Objects: PyRASA provides functions designed for both continuous (Raw) and event-related (Epochs) data, making it versatile for various types of EEG/MEG analyses.
Consistent Ontology: PyRASA uses the same jargon to label parameters as specparam, the most commonly used tool to parametrize power spectra, to allow users to easily switch between tools depending on their needs, while keeping the labeling of features consistent.
Custom Aperiodic Fit Models: In addition to the built-in “fixed” and “knee” models for aperiodic fitting, users can specify their custom aperiodic fit functions, offering flexibility in how aperiodic components are modeled.
Example Usage#
Decompose spectra into periodic and aperiodic components:
from pyrasa.irasa import irasa
irasa_out = irasa(sig,
fs=fs,
band=(.1, 200),
nperseg=duration*fs,
noverlap=duration*fs*overlap,
hset_info=(1, 2, 0.05))
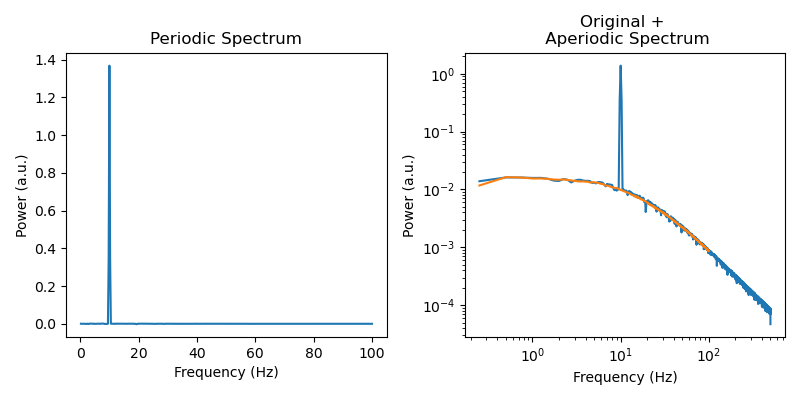
Extract periodic parameters:
irasa_out.get_peaks()
ch_name |
cf |
bw |
pw |
|---|---|---|---|
0 |
10.0 | 1.1887 | 0.495 |
||
Extract aperiodic parameters:
irasa_out.fit_aperiodic_model(fit_func='knee').aperiodic_params
Offset |
Knee |
Exponent_1 |
Exponent_2 |
fit_type |
Knee Frequency (Hz) |
tau |
ch_name |
|---|---|---|---|---|---|---|---|
4.329e-17 |
62.11 |
0.0552 |
1.4602 |
knee |
13.855 |
0.0115 |
0 |
And the goodness of fit:
irasa_out.fit_aperiodic_model(fit_func='knee').gof
mse |
r_squared |
BIC |
AIC |
fit_type |
ch_name |
|---|---|---|---|---|---|
0.000081 |
0.999303 |
-31.9892 |
-47.9550 |
knee |
0 |
If you want to reproduce the example above checkout this example notebook.
How to Contribute#
Contributions to PyRASA are welcome! Whether it’s raising issues, improving documentation, fixing bugs, or adding new features, your help is appreciated.
To file bug reports and/or ask questions about this project, please use the Github issue tracker.
Please refer to the CONTRIBUTING.md file for more information on how to get involved.
Reference#
If you are using IRASA, please cite the smart people who came up with the algorithm:
Wen, H., & Liu, Z. (2016). Separating fractal and oscillatory components in the power spectrum of neurophysiological signal. Brain Topography, 29, 13-26. https://doi.org/10.1007/s10548-015-0448-0
If you are using PyRASA, it would be nice if you could additionally cite us (whenever the paper is finally ready):
Schmidt F., Hartmann T., & Weisz, N. (2025). PyRASA - Spectral parameterization in python based on IRASA.