Improving your IRASA model fits#
When fitting IRASA we sometimes face the situation that residual periodic activity remains in our aperiodic spectrum. In this notebook I will illustrate two possible ways that could lead to this situation and how to avoid it. First we start again by simulating a fairly simple signal consisting of a 10Hz oscillation and an aperiodic exponent of 1.
[1]:
from neurodsp.sim import sim_combined
from neurodsp.utils import create_times
import matplotlib.pyplot as plt
from pyrasa.irasa import irasa
[2]:
fs = 500
n_seconds = 60
sim_components = {'sim_powerlaw': {'exponent' : -1},
'sim_oscillation': {'freq' : 10}}
sig = sim_combined(n_seconds=n_seconds, fs=fs, components=sim_components)
times = create_times(n_seconds=n_seconds, fs=fs)
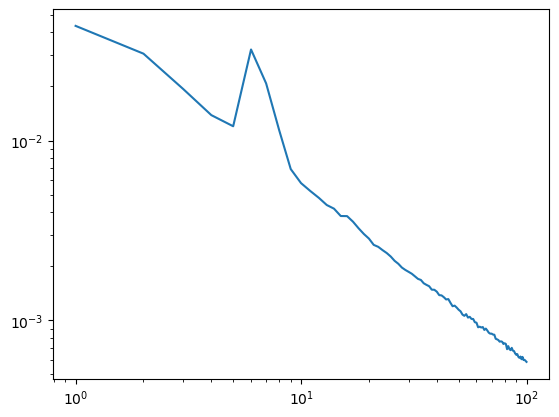
Case #1: Increase your frequency resolution#
When the frequency resolution for your psd is “low” up/-downsampling is not enough to reliably get rid of peaks in the signal. Increasing the length of each segment (i.e. the “duration”) should help you out.
[3]:
#low segment length
duration=1
overlap=0.5
irasa_out = irasa(sig,
fs=fs,
band=(1, 100),
nperseg=int(duration*fs),
noverlap=int(duration*fs*overlap),
hset_info=(1, 2, 0.05))
plt.loglog(irasa_out.freqs, irasa_out.aperiodic.T)
irasa_out.fit_aperiodic_model().aperiodic_params
[3]:
| Offset | Exponent | fit_type | ch_name | |
|---|---|---|---|---|
| 0 | -1.355433 | 1.053051 | fixed | 0 |
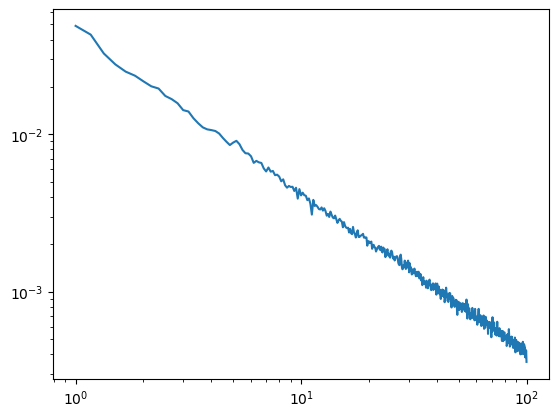
[48]:
#Increased segment length
duration=6
overlap=0.5
irasa_out = irasa(sig,
fs=fs,
band=(1, 100),
psd_kwargs={'nperseg': int(duration*fs),
'noverlap': int(duration*fs*overlap),
},
hset_info=(1, 2, 0.05))
plt.loglog(irasa_out.freqs, irasa_out.aperiodic.T)
irasa_out.fit_aperiodic_model().aperiodic_params
[48]:
| Offset | Exponent | fit_type | ch_name | |
|---|---|---|---|---|
| 0 | -1.361501 | 1.012603 | fixed | 0 |
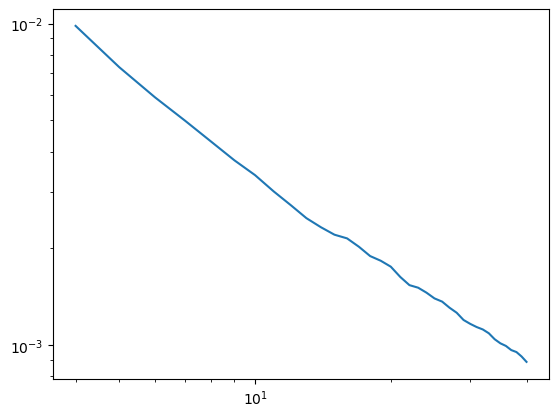
Case #2: Increase your hmax#
Sometimes we either can’t or don’t want to increase our frequency resolution. In this case we can increase our maximal up/-downsampling factors to get rid of peaks in the signal. However, be aware that this can - depending on our filter settings and sampling rate - limit the frequency range that we can sensibly evaluate (see irasa_pitfalls.ipynb).
[4]:
#low segment length, but high hset
duration=1
overlap=0.5
irasa_out = irasa(sig,
fs=fs,
band=(4, 40),
nperseg=int(duration*fs),
noverlap=int(duration*fs*overlap),
hset_info=(1.1, 6, 0.05))
plt.loglog(irasa_out.freqs, irasa_out.aperiodic.T)
irasa_out.fit_aperiodic_model().aperiodic_params
[4]:
| Offset | Exponent | fit_type | ch_name | |
|---|---|---|---|---|
| 0 | -1.464617 | 0.994558 | fixed | 0 |
[ ]: